Research Focus
Our lab investigates how the cell cycle is integrated with developmental and physiological processes in multicellular organisms. Disruption of this coordination can lead to diseases such as cancer, tissue degeneration, and ageing.
We focus on a group of evolutionarily conserved proteins known as Cell Cycle Regulators (CCRs). These proteins, traditionally recognised for their roles in cell proliferation, are increasingly appreciated for their additional functions in regulating cell fate and differentiation. We previously proposed that these proteins serve as intracellular coordinators of proliferation and differentiation (Kimata et al., 2020).
To study these novel functions, we use Drosophila melanogaster as our main model organism, combining in vivo genetics, high-resolution imaging, multi-omics analysis, and tailored biochemical approaches. We also translate our findings into human biology by studying conserved mechanisms in cultured human cells and mammalian organoids. Our goal is to uncover the molecular logic that synchronises cell cycle control with development and tissue function across evolution. Ultimately, we hope our work will inform new therapeutic strategies for currently incurable diseases.
We welcome collaborations and enthusiastic students to join our current projects:
1. Non-Canonical Functions of Cell Cycle Regulators in Cell Fate Control
Recent work, including ours, has shown that CCRs can directly influence developmental signals and transcriptional programmes (Kimata, 2019, Kimata, et al., 2020). For example, the APC/C, a master regulator of the cell cycle, suppresses the Wnt pathway by degrading the positive modulator Nek2, thus linking cell cycle exit to differentiation in the developing Drosophila eye (Martins et al., 2017).
We expanded this concept by conducting a genome-wide screen of CCRs to uncover novel, cell cycle-independent roles in Drosophila. We identified multiple CCRs that are essential for retinal differentiation, including CDKs, and are now investigating their molecular mechanisms (Man, Ding, et al., unpublished). To explore evolutionary conservation, we are also studying mammalian orthologs of these CCRs in human cell cultures, organoids, and mouse models.
The genome-wide screen for cell cycle-independent functions of CCRs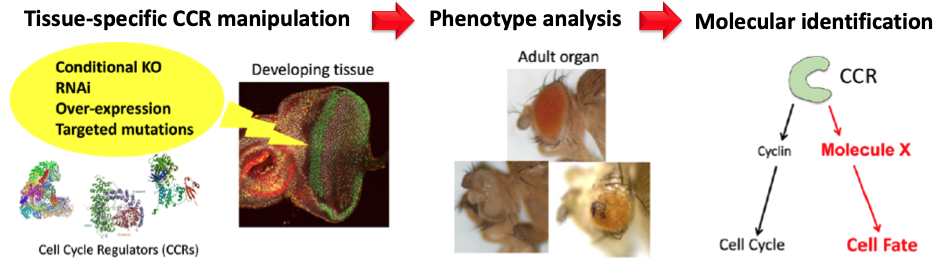
2. The Regulation of Cellular Quiescence and G0 Phase
Many adult cells enter non-dividing, dormant states. Tissue stem cells often enter reversible quiescence (G0 phase), while fully differentiated cells such as neurons exit the cell cycle permanently. Despite their significance, the molecular mechanisms that control these states remain poorly understood.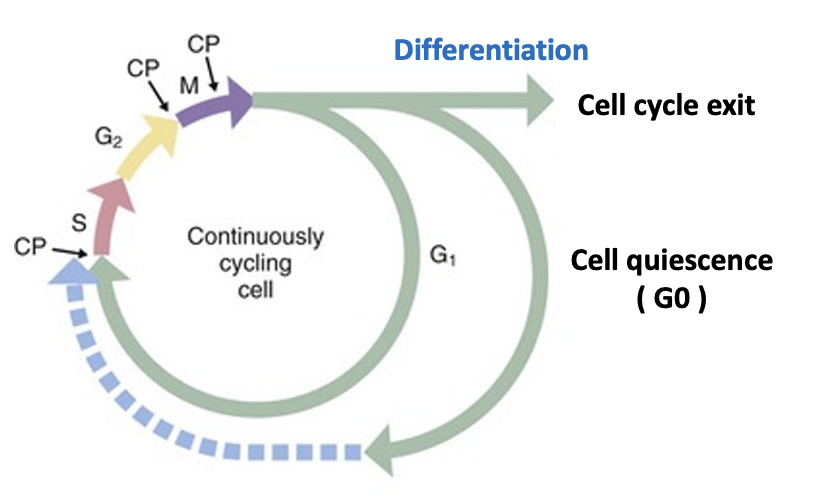
We study these quiescent states both in vivo and in vitro. In Drosophila, we investigate quiescence in adult brain and gut tissues, focusing on its regulation and impact on tissue homeostasis (Chen, Man, Shu, et al., 2025, bioRxiv). In parallel, we use cultured human cells and rapid protein-targeting systems to dissect the coordination between G0 entry and genomic states, aiming to clarify how quiescence contributes to health and disease.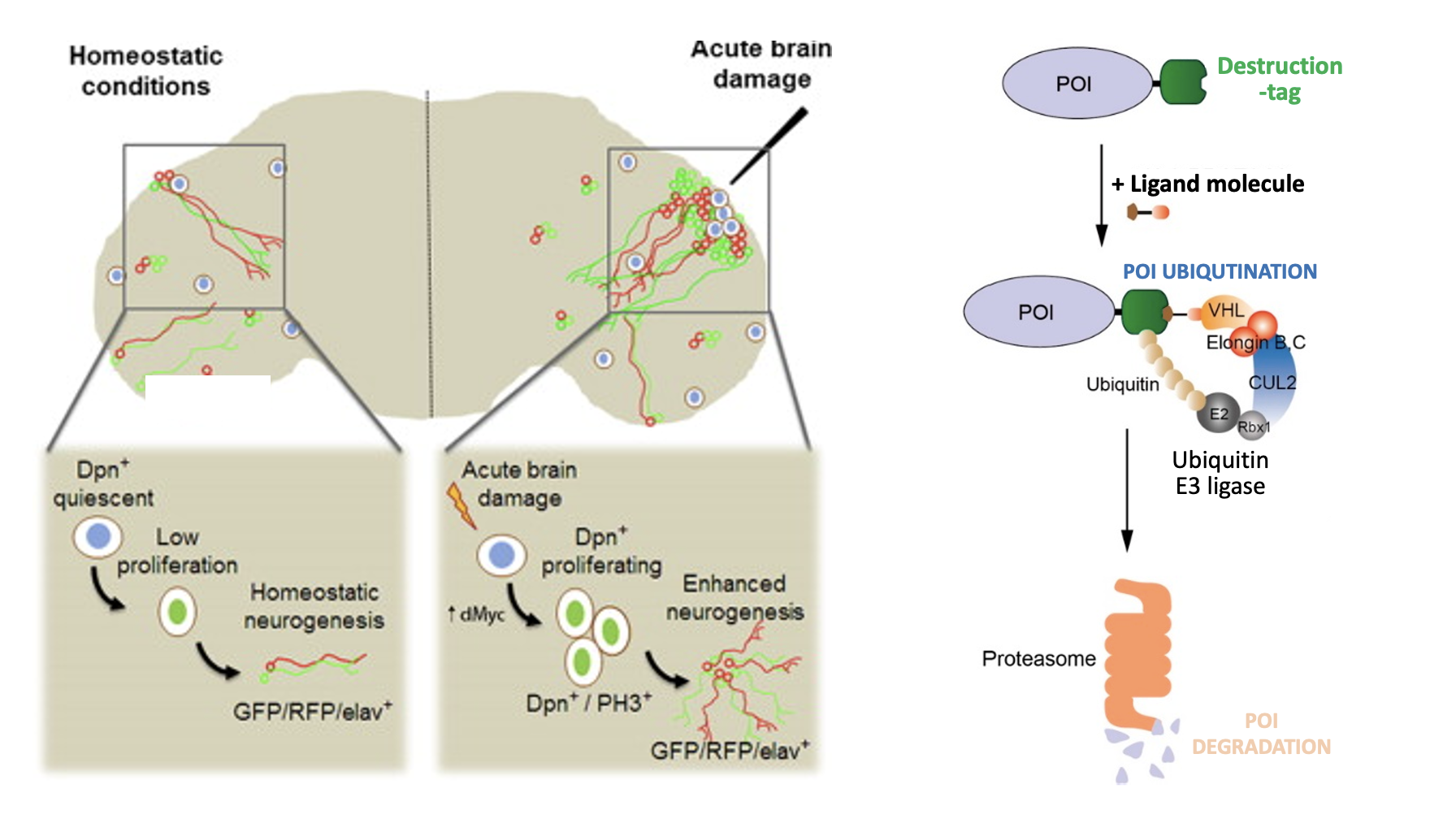
3. Centrosomes, Cilia, and Cell Cycle Regulators in Development and Disease
The centrosome orchestrates cytoskeletal dynamics, and in non-dividing cells, it can become the primary cilium—a signalling hub. Dysfunction of centrosomes and cilia is linked to diseases ranging from cancer to ciliopathies.
Multiple CCRs interact with centrosomes and cilia. In collaboration with labs at Cambridge and Institut Curie, we showed that APC/C’s co-activator CDH1 (Fzr in Drosophila) binds Spd2/CEP192 to regulate Aurora A degradation at centrosomes, affecting neural stem cell maintenance (Meghini et al., 2016; 2023). We also demonstrated its role in asymmetric division and centrosome asymmetry (Gambarotto et al., 2019).
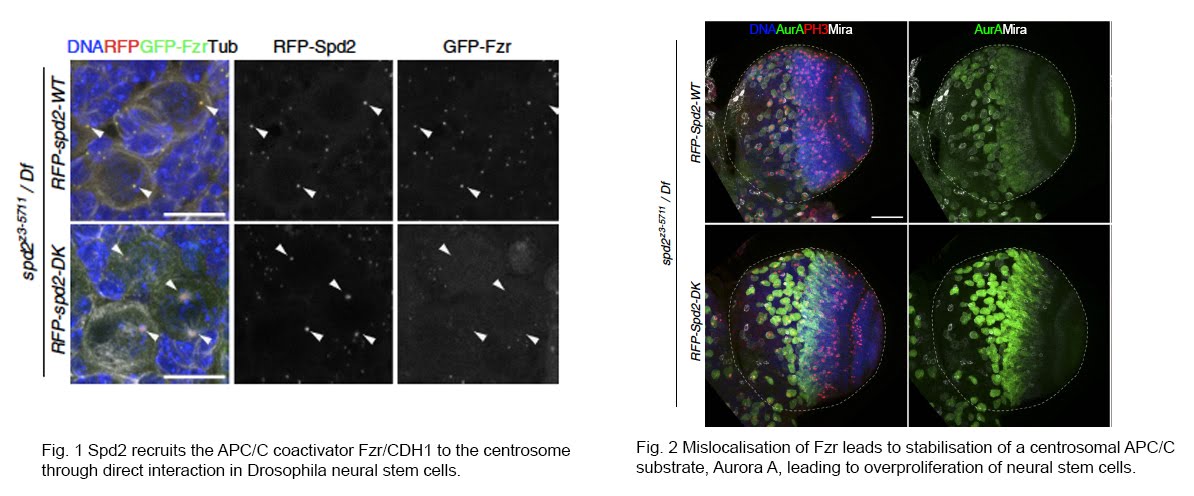
Using CRISPR-based mutations in the APC/C cofactor Vihar/Ube2C, we discovered a role for APC/C in regulating centrosome migration during oocyte specification via degradation of Polo kinase (Braun et al., 2021)
We are continuing to explore the interactions between CCRs and centrosome/cilia components and their functions in various Drosophila tissues and human cells and organoids.
Recent publications
- , , , , , ,
- Meghini F, Martins T, Zhang Q, et al., APC/C-dependent degradation of Spd2 regulates centrosome asymmetry in Drosophila neural stem cells. EMBO Rep. 2023 April.
- Braun AL, Meghini F, Villa-Fombuena G, et al., The careful control of Polo kinase by APC/C-Ube2C ensures the intercellular transport of germline centrosomes during Drosophila oogenesis. Open Biol. 2021 June.
- Kimata Y, Leturcq M, Aradhya R. Emerging roles of metazoan cell cycle regulators as coordinators of the cell cycle and differentaition. FEBS Letters. 2020 July.
- Ochi T, Quarantotti V, Lin H, et al. CCDC61/VFL3 is a paralog of SAS6 and promotes ciliary functions. Structure. 2020 June.
- Gambarotto D, Pennetrier C, Rynaiawec JM, et al. Plk4 regulates centriole asymmetry and spindle orientation in neural stem cells. Developmental Cell. 2019 Jul.
- Kimata Y. APC/C Ubiquitin Ligase: Coupling Cellular Differentiation to G1/G0 Phase in Multicellular Systems. Trends in Cell Biology. Apr. 2019
- Martins T, Meghini F, Florio F, Kimata Y. The APC/C coordinates retinal differentiation with G1 arrest through the Nek2-dependent modulation of Wingless signalling. Developmental Cell. 2017 Jan.
- Meghini F, Martins T, Tait X (equally contributed), Fujimitsu K, Yamano H, Glover DM, Kimata Y. Targeting of Fzr/Cdh1 for timely activation of the APC/C at the centrosome during mitotic exit. Nature Comms. 2016 Aug 25;7:12607.
